
The medical test battling antibiotic resistance
Antibiotic resistance is one of the biggest global health threats we face. As the problem grows, we’re running the risk of bacterial infections becoming fiendishly difficult to treat. This in turn makes routine medical treatments such as surgeries and chemotherapy far riskier.
Overprescription is a major contributor to antibiotic resistance. This can happen when GPs can’t diagnose the exact bacteria causing common infections. Confirmation usually involves sending a sample to be analysed in a microbiology lab, which can take several days. Patients can’t always wait this long for treatment.
So, to avoid severe complications such as sepsis, doctors must sometimes prescribe antibiotics before they know if they’ll be effective. The problem is, whenever antibiotics are used, bacteria can become resistant to them.
Now, a Swedish spinout company, Sysmex Astrego, has developed a test that can determine whether (and which) antibiotics should be prescribed to treat a urinary tract infection in a matter of minutes. It shrinks the suite of tests that normally require a microbiology lab into a compact machine designed for GP’s offices and A&E departments. The test won the Longitude Prize on antimicrobial resistance in 2024 run by Challenge Works.
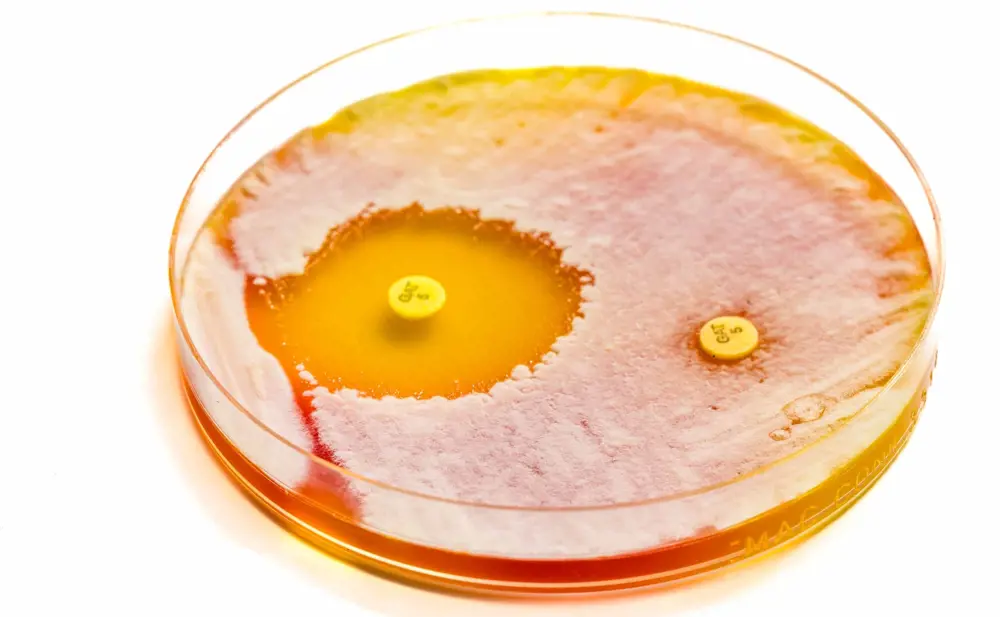
Medical microbiologists grow bacteria sampled from a patient’s infection in a dish and expose them to different antibiotics. If the bacteria die (left hand side), doctors will be able to successfully treat the patient with the antibiotic. If the bacteria survive (right hand side), they are resistant. The process takes several days © Shutterstock
The test requires just 400 microlitres of urine, which is injected into a cartridge the size of a smartphone. Air pressure is used to manoeuvre the sample through tiny channels and into traps that capture the bacteria, holding them in view of a microscope at a high magnification while feeding them with antibiotics. Every minute, it takes about 60 images of the channels to see whether bacteria are present. If they are, it calculates how quickly the bacteria grow with different antibiotics, or with no antibiotics.
“You can clearly see if an antibiotic starts slowing down the growth rate,” says Mikael Olsson, the company’s CEO. For some antibiotics, he adds, you see the bugs “pop like balloons” in the fluidic traps.
For some antibiotics, Olsson adds, you see the bugs “pop like balloons” in the fluidic traps.

For Sysmex Astrego’s test, a urine sample of less than half a millilitre is loaded into a smartphone-sized cartridge. The cartridge is then inserted into an analyser, which exposes bacteria to different antibiotics and provides results in minutes © Sysmex Astrego
Antibiotic testing on a chip
The test’s beginnings go back to 2015, when Swedish biophysicist Professor Johan Elf was developing ways to study chemical reactions inside individual bacteria at Uppsala University. This involved channelling liquids containing bacteria down tiny tubes and looking at them under a microscope.
These tiny tubes are called microfluidic and nanofluidic traps. They’re sometimes called lab-on-a-chip technologies, as their precisely patterned channels are embedded in small silicone and glass chips. At their largest, the channels are hundreds of micrometres wide. Nanofluidics, on the other hand, contain channels narrower than one micrometre wide – about a hundredth of the width of a human hair. At this scale, the properties of fluids change. “You get very good control over what goes where and when,” says Elf. “You can spatially and temporally time when different chemistries occur.”
With this precise control over different fluids, Elf and his team could pump in different antibiotics and bacteria to see if the microbes responded to the drugs or not. “The first time I saw that the bacteria died in front of our eyes in just a couple of minutes – that was really a critical moment to see this will actually work,” says Elf. “It’s unusual you get that clear-cut results in the lab.”
Once he realised its potential usefulness, Elf patented the technology and spun out a company, with plans to make the incredibly complex and expensive system cheaper and more user-friendly.
Squeezing a lab setup into the size of a printer
Turning what was at the time a biophysics lab setup, custom-built for studying cells, into something more or less the size of a desktop printer, was a major challenge. It took two years to develop a prototype. Then, it had to be robust and easy to manufacture, with software and data analysis challenges to address along the way.
The second big challenge came when the device’s first clinical tests came along. Urine samples vary enormously between patients. They may or may not contain bacteria. Those bacteria may or may not die when they’re exposed to antibiotics. Other factors change how the machine reads any one sample such as the cocktail of various human cells it contains.
The fluidic technology helped. Big cells tend to stay away from surfaces, whereas small cells move closer to surfaces, explains Elf. In practice, this means the test can filter out most of the types of cell that you don’t want to analyse. Urine samples, for instance, contain red blood cells, white blood cells, and a variety of other cells other than bacteria.
Olsson explains the team found a way to compensate for the different parameters, by collecting clinical samples showing the full range of how bacteria might respond to antibiotics, from the vulnerable to the very resistant ‘superbugs’.
Bringing back discarded antibiotics
While antimicrobial resistance is still growing, access to precise and rapid diagnostics will allow us to use the antibiotics we have available.
“We can bring back antibiotics that have been discarded because of the high resistance [to them],” says Elf. Even if 20% of the population are resistant, he adds, we can still use them for the other 80% of people.
With proper diagnostics, we can also opt for more narrow-spectrum antibiotics, rather than settling for broad spectrum antibiotics that drive resistance.
Following the 10-year journey that led them to winning the Longitude Prize, Olsson explains the company is now ramping up production to meet demand in Europe and hopes to expand to the US and Asia.
The next stop after that could be applying the diagnostic tool to new infections. Among the candidates are sepsis, lower respiratory tract infections, and tuberculosis. Elf has already begun work on tuberculosis at Uppsala University, which currently takes four weeks to diagnose. He hopes to push this down to 12 hours, although acknowledges that it will take time until there is a product for tuberculosis. “There are lots of different challenges in terms of the microfluidics because they grow in a different way,” he explains. “The beauty of this platform is that it’s quite applicable to lots of different infections,” says Olsson. “It’s a smorgas-table of selecting the next one.”
Contributors
Florence Downs
Author
Keep up-to-date with Ingenia for free
SubscribeRelated content
Health & medical
A gamechanger in retinal scanning
2006 MacRobert Award winner Optos rapidly became a leading medical technology company and its scanners have taken millions of retinal images worldwide. There is even a display at the Science Museum featuring the Optos development. Alastair Atkinson, of the award-winning team, describes the personal tragedy that was the trigger for the creation of Optos.

Kidney dialysis
Small haemodialysis machines have been developed that will allow more people to treat themselves at home. The SC+ system that has been developed is lighter, smaller and easier to use than existing machines.

Engineering polymath wins major award
The 2015 Queen Elizabeth Prize for Engineering has been awarded to the ground-breaking chemical engineer Dr Robert Langer FREng for his revolutionary advances and leadership in engineering at the interface between chemistry and medicine.

Blast mitigation and injury treatment
The Royal British Legion Centre for Blast Injury Studies is a world-renowned research facility based at Imperial College London. Its director, Professor Anthony Bull FREng, explains how a multidisciplinary team is helping protect, treat and rehabilitate people who are exposed to explosive forces.
Other content from Ingenia
Quick read

- Environment & sustainability
- Opinion
A young engineer’s perspective on the good, the bad and the ugly of COP27

- Environment & sustainability
- Issue 95
How do we pay for net zero technologies?
Quick read

- Transport
- Mechanical
- How I got here
Electrifying trains and STEMAZING outreach

- Civil & structural
- Environment & sustainability
- Issue 95